Laboratory Products
Bio Workbench Enables Scientists to Achieve New Breakthroughs in Biosciences
Apr 15 2010
NVIDIA announced the Tesla Bio Workbench, which enables scientists to push the boundaries of biological research by turning a standard PC into a ‘computational laboratory’ capable of running complex bioscience codes in fields such as drug discovery and DNA sequencing more than 10-20 times faster through the use NVIDIA® Tesla™ GPUs.
The Tesla Bio Workbench consists of: a range of GPU-optimised bioscience applications for molecular dynamics - and quantum chemistry-based research, including: AMBER, GROMACS, LAMMPS, NAMD, TeraChem, VMD, and bioinformatics applications like CUDASW++ (Smith-Waterman), GPU-HMMER, and MUMmerGPU; a community site for downloading the applications, checking out the latest benchmarks, reading academic papers and tutorials, joining discussion forums with the application developers themselves and more; and details on the Tesla GPU-based workstations and clusters available worldwide for easy deployment of these applications.
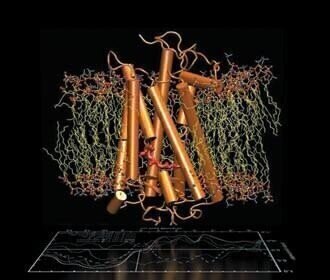
Scientists have traditionally performed experiments in laboratories, where chemicals are combined, their interactions studied and their effectiveness measured. Advances in computational science have now enabled these experiments to be carried out using molecular dynamics and quantum chemistry simulation models, however these have typically required very large supercomputers with thousands of central processing units (CPUs). By using the massively parallel CUDA™ architecture of NVIDIA GPUs, these applications can now be run 10-20 times faster, which means even a PC with Tesla GPUs can outperform a supercomputer.
"We are working on a new GPU-based technique in the VMD molecular dynamics visualisation software that investigates how small molecules like oxygen and CO2 migrate inside proteins. This research is critical in the study of enzymatic reaction mechanisms," said John Stone, Senior Research Programmer, University of Illinois at Urbana-Champaign. “A simulation that takes one day to run on a GPU-based workstation would have taken 30 days to run on a CPU-based machine, rendering it impractical for real research.”
Digital Edition
Lab Asia 31.2 April 2024
April 2024
In This Edition Chromatography Articles - Approaches to troubleshooting an SPE method for the analysis of oligonucleotides (pt i) - High-precision liquid flow processes demand full fluidic c...
View all digital editions
Events
May 14 2024 Oklahoma City, OK, USA
May 15 2024 Birmingham, UK
May 21 2024 Lagos, Nigeria
May 22 2024 Basel, Switzerland
Scientific Laboratory Show & Conference 2024
May 22 2024 Nottingham, UK









.jpg)





