-
 Prof Jim Naismith and Dr Lucile Moynie. Credit Rosalind Franklin Institute
Prof Jim Naismith and Dr Lucile Moynie. Credit Rosalind Franklin Institute -
-
 Professor Ray Owens. Credit Rosalind Franklin Institute
Professor Ray Owens. Credit Rosalind Franklin Institute
News & Views
Engineered Llama Antibodies Neutralise Covid-19 Virus
Jul 17 2020
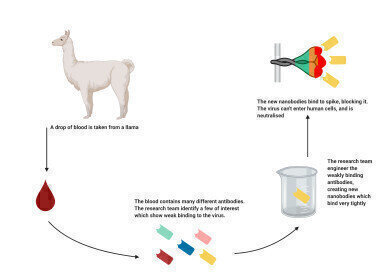
Researchers have developed a family of engineered nanobodies which neutralise the SARS-CoV-2 virus, targeting the viral spike protein in a novel way.
The research team from The Rosalind Franklin Institute, working with colleagues at Diamond Light Source, Oxford University and Public Health England, have hailed the breakthrough as a potential therapy which could be delivered as part of a combination in a synthetic convalescent serum. The nanobodies could also be developed as a powerful diagnostic, taking advantage of their very high specificity and affinity.
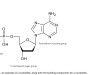
The family of nanobodies were engineered from a library of antibodies derived from llamas. These nanobodies bind very tightly (around 10 nM) to the spike protein of the SARS-CoV-2 virus, blocking it from binding to human cells and preventing infection. Importantly the exceptionally high affinity is preserved for multiple conformations of the spike.
Tests on live virus, carried out in specialist labs in Oxford and Porton Down, show that the nanobodies neutralise SARS-CoV-2, preventing it from infecting cells. The neutralisation concentration was below 10 nM marking out these agents as extremely potent. Most importantly the lab work shows that one of the nanobodies, H11-H4, can be combined with a human antibody to give an additive effect, which has two advantages. Firstly, the combination is more powerful than either component alone. Secondly, combinations are very valuable because they dramatically reduce the risk of the virus mutating to escape.
There is currently no 'cure' or vaccine for COVID-19. However, transfusion of critically ill COVID-19 patients with serum from convalesced individuals has been shown to improve clinical outcomes. This would suggest that neutralisation of the virus using antibodies could be a useful therapy.
Professor James Naismith, Director of The Rosalind Franklin Institute and Professor of Structural Biology at Oxford University said: “With this work, a world leading team of scientists created, analysed and tested the nanobody in 12 weeks. We have carried out experiments in just a few days that would typically take months to complete. This was only possible because so many scientists set aside their other work and brought their skills to bear. The ability to work at speed and with purpose is essential in an unfolding pandemic, and we are hopeful that we can push this breakthrough on into pre-clinical trials.”
Using advanced imaging with X-rays and electrons at Diamond Light Source, the team can see that the nanobodies bind to the spike protein in a new and different way to other antibodies already discovered. These insights allowed the team to identify the human antibody to combine with H11-H4 to create the potent combination.
Professor David Stuart, from Diamond Light Source and Oxford University said “The UK has cutting edge structural biology facilities that allow us to see the atoms in molecules. This drives the research to treat Covid19 forward at great pace. Diamond and Oxford collaborated to ensure immediate structural insights for this important work. We discovered that the nanobodies bind to the spike, covering up the portion that the virus uses to enter cells.”
Professor Ray Owens from Oxford University, who leads the nanobody program at the Franklin, said: “I think we have shown the value of building the advanced capability for nanobody discovery in the Franklin. We went from a standing start to powerful unique molecules in just a few weeks. This rapid response will be applicable to other epidemics which emerge in the future, and to other urgent research problems.”
The Franklin works in partnership with the University of Reading, whose herd of llamas have been providing antibodies to the team. The immune system of llamas, camels and alpacas naturally produce heavy chain only antibodies, so without a partner light chain. From these antibodies, researchers can derive single domain binding agents, known as nanobodies, that are very powerful tools to stabilise protein conformation for molecular imaging.
Initially, the Franklin team started with a lab-based approach – isolating nanbodies from existing libraries and engineering them to produce optimum binding which has enabled a very rapid turnaround. The team are now working on the next generation of nanobodies, engineered from natural antibodies created following the immunisation of a llama at the University Reading with harmless purified proteins from the virus. Already nanobodies that bind differently to those already identified have been produced, opening up opportunities for testing potentially powerful combinations of nanobodies for neutralising the SARS-CoV-2 virus.
Digital Edition
Lab Asia 31.2 April 2024
April 2024
In This Edition Chromatography Articles - Approaches to troubleshooting an SPE method for the analysis of oligonucleotides (pt i) - High-precision liquid flow processes demand full fluidic c...
View all digital editions
Events
Apr 22 2024 Marrakech, Morroco
Making Pharmaceuticals Exhibition & Conference
Apr 23 2024 Coventry, UK
Apr 23 2024 Kintex, South Korea
Apr 23 2024 Seoul, South Korea
Apr 24 2024 Jakarta, Indonesia







.jpg)